Phylogeography of mitochondrial DNA in West Eurasia
Today’s West Eurasian genetic population structure is the result of several migration events that originated somewhere between East Africa and the Persian Gulf ~45,000 years ago [Torroni 2006]. Two major movements of humans took place during the Upper Palaeolithic before and after the Last Glacial Maximum (LGM). The Franco-Cantabrian refuge area of south-western Europe was the source of a late-glacial expansion of hunter-gatherers that repopulated northern Europe after the LGM [Achilli 2004].
More than three-quarters of the present-day European mtDNA gene pool most likely comes from indigenous Mesolithic or Palaeolithic ancestors. In addition Neolithic immigrants, who brought the first forms of agriculture to the hunter-gatherer dominated European landscape, ~8,000 years ago play an important part of today’s mtDNA pool of modern humans in West Eurasia [Richards 2000].
According to our current knowledge all non-Africans descend from two haplogroups (M and N) that derived from the African haplogroup L3. One branch (M) moved to East Asia, whereas the sister clade N spread into the Near East and Europe subsequently. Haplogroup N diverged further to give rise to haplogroup R, which is the founder of most West-Eurasian mtDNA lineages. Three R branches (haplogroups R0, JT and U) and additionally three minor N branches (N1, N2, X) are found in the present European mtDNA pool. Thus Europeans and Near Easterners share a rather recent common ancestry, since they share essentially the same set of haplogroups. Nearly half of the West Eurasian assemblage of human mtDNA is fractioned into numerous sub-lineages of the predominant haplogroup R0 (R0a, HV). Subhaplogroup H, descending from R0, is dominating with a considerable proportion of 40%-50% of all mtDNAs and can be further dissected into numerous sub-haplogroups.
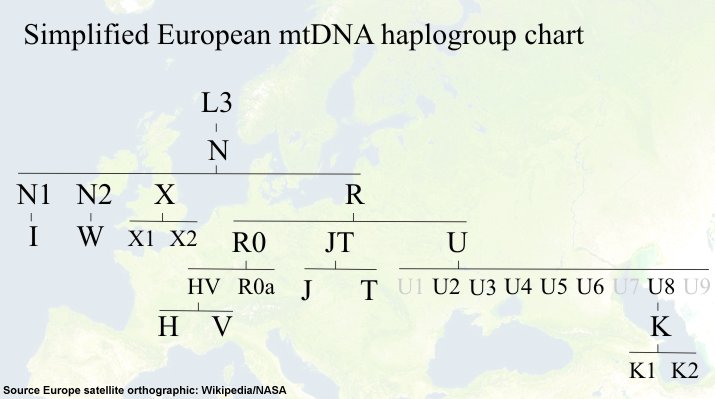
Haplogroup V, a sister clade of H, can be found in Northern Scandinavia at a very high ratio (~60%) [Kivisild 2003]. Haplogroup U has an extremely broad geographic distribution that ranges from Europe and North Africa to India and Central Asia and has a very high overall frequency (15%-30%). This haplogroup arose shortly after the “out of Africa” exit and rapidly radiated into numerous distinct subclades (U1-U9) [Achilli 2005]. One of this subclades, haplogroup K (derived from haplogroup U8) is present all over Europe, but very prominent in Ashkenazi populations [Behar 2006]. One famous European, the “iceman” from the Austro-Italian Alps is a member of haplogroup K. Haplogroup J shows a greater genetic variability in the Middle East compared to Europe. Haplogroup T is present in about 20% of the mtDNA pool in West Eurasia. Also here we know one intriguing haplogroup T member Tsar Nicholas II of Russia.
To understand the distribution of present-day’s European haplogroups and provide a basis for frequency estimation European mtDNA haplotypes are analysed in population studies and stored and provided to the community in EMPOP.
Literature cited
Torroni A 2006 Trends Genet 22: 339-345
Achilli A 2004 Am J Hum Genet 75: 910-918
Richards M 2000 Am J Hum Genet 67: 1251-1276
Ingman M 2007 Eur J Hum Genet 15: 115-120
Kivisild T 2003 Am J Hum Genet 72: 313-332
Quintana-Murci L 2004 Am J Hum Genet 74: 827-845
Achilli A 2005 Am J Hum Genet 76: 883-886
Behar DM 2006 Am J Hum Genet 78: 487-497