Phylogeography of mitochondrial DNA in Africa
Background and state of research
Today, it is a generally accepted fact that the cradle of humanity and the main location of anatomically modern humans for most of their existence is in the African continent. Nevertheless, due to the lack of sufficient archaeological findings, the initial Homo sapiens population dynamics, dispersal-, and migration routes remain poorly understood [Cann 1987, Underhill 2007]. The potential to use present day genetic patterns to detect the existence, or lack thereof, of matrilineal genetic structure among early Homo sapiens populations in sub-Saharan Africa is therefore of particular interest [Behar 2008]. Based on the information of recent mtDNA sampling, main population structures were attempted to be identified and major potential migration-, and dispersal routes reconstructed [Behar 2008, Salas 2002].
The human mtDNA phylogeny can be collapsed into two daughter branches, L0 and L1’2’3’4’5’6 located on opposite sides of its root [van Oven 2008]. The L1’2’3’4’5’6 branch is wide spread all over the African continent and gave rise to almost every existing haplogroup today. Recent works have been supporting the out–of-Africa scenario 50,000 - 65,000 years before present confirming the moving out of some L3 members to the Arabian region. Diverging clades of L3, N and M respectively, then colonized the remaining continents in a way that the M lineages moved to East Asia passing the costal line of India whereas the N lineages either spread in the Arabian region or moved to and diverged within Europe [Mellars 2006, Macaulay 2005, Torroni 2006].
The mitochondrial gene pool in Africa is diverse due to the long history of dispersals and migrations within the last ~150,000 years until the beginning of permanent settlements ~18,000 years before present. The most recent widespread demographic shift within the continent was most probably the Bantu dispersal, with archaeological and linguistic evidence suggesting an origin in West Africa 3,000–4,000 years ago, spreading both east and south [Behar 2008]. Hence, the present genetic population structure is composed of original populations overlaid by populations resulting from demographic shifts.
However, some regional accumulations of specific haplogroups can be outlined.
The North of Africa is characterized by the presence of non-African haplogroups M1 and U6 being consequences of back migrations from the Arabian peninsula. Haplogroup L1c is accumulated in Central Africa as compared to the remaining regions. Haplogroups L0d and L0k are prevalent solely in South Africa. The main dispersal of L0a haplogroups ranges from the Northeast to the Southeast reflecting a possible migration rout.
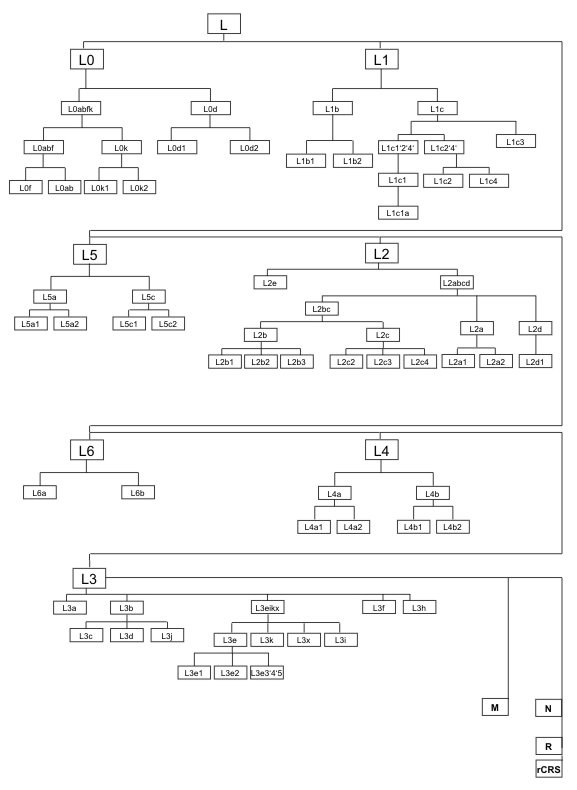
Figure 1: Overview of the African phylogeny. The African macro-haplogroup L is subdivided into the haplogroups L0, L1, L5, L2, L6, L4, and the youngest L3, that comprises also all non-African lineages. This figure represents a condensed phylogeny and does not comprise all known haplogroups within macro-haplogroup L.
Aims
100,000 years of African population structure, migrations and dispersals are still poorly understood. With our investigations on selected African populations we will contribute to shed more light on the African phylogeny. The African sequences will be incorporated to the EMPOP database thus contributing to gain a representative high quality worldwide dataset of mtDNA sequences.
Literature cited
Cann RL 1987 Nature 325:31-36
Underhill PA 2007 Annu Rev Genet 41:539-564
Behar DM 2008 Am J Hum Genet 82:1130-1140
Salas A 2002 Am J Hum Genet 71:1082-1111
van Oven M 2008 Hum Mutat 1039:e386-394
Mellars P 2006 Science 313:796-800
Macaulay V 2005 Science 308:1034-1036
Torroni A 2006 Trends Genet 22:339-345